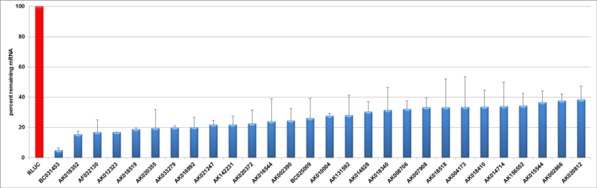
esiRNA non-coding mouse
The esiRNA technology is a powerful tool for loss of function gene studies. To illustrate the potency of silencing triggers quantitative real time (qRT)-PCR is often used to measure the knockdown rates. The knockdown validation on mRNA level was here performed 24 hours post transfection of esiRNA using qRT-PCR. To be able to assess knockdown rates, the expression levels of the mRNA of interest was compared to cells (HeLa or mouse ES) simultaneously transfected with Renilla Luciferase (negative control).
However, a more valuable measure of the knockdown potency in an RNAi experiment is the reduction in protein level. We therefore also validate the knockdown rates of esiRNAs on protein level using quantitative western blot analysis (Odyssey, Li-COR). The time point for maximum knockdown rate for each protein can vary significantly, as it is depending on factors such as protein stability, turnover rate or cell proliferation rates. The knockdown validation on protein level was here performed at 72 hours post transfection of esiRNA in HeLa cells. To be able to assess knockdown rates, the expression levels of the protein of interest was compared to HeLa cells simultaneously transfected with Renilla Luciferase (negative control).
Two approaches were used to monitor knockdown at protein level: 1. Specific antibodies for the protein of interest were used for the quantitative western blot analysis. 2. Proteins of interest were GFP-tagged on a bacterial artificial chromosome (BAC) and stably integrated into the genome of HeLa cells, allowing for near physiological expression (Poser I. et al Nat Methods. 2008 May;5(5):409-15). Using a GFP antibody the detection by quantitative western blot analysis of the protein of interest is straightforward.
* Average of 2 technical replicates
| esiRNA ID | Accession No. | LNC RNA Name | LNC RNA Description | Ensembl ID | RefSeq ID | Knock-Down Rate | |
| MNC-00089-1 | BC031453 | NR_046233.1 | 95,05 ± 1,66 | ||||
| MNC-00018-1 | AK018352 | 84,59 ± 2,18 | |||||
| MNC-00010-1 | AF032130 | 83,26 ± 8,18 | |||||
| MNC-00060-1 | AK012323 | 83,11 ± 0,08 | |||||
| MNC-00063-1 | AK018519 | 83,31 ± 0,91 | |||||
| MNC-00087-1 | AK020355 | 80,5 ± 12,19 | |||||
| MNC-00173-1 | AK033279 | 80,1 ± 1,17 | |||||
| MNC-00492-1 | AK016992 | 80 ± 6,6 | |||||
| MNC-00102-1 | AK021247 | C430039J16Rik | RIKEN cDNA C430039J16 gene | ENSMUSG00000091451 | 78,28 ± 2,97 | ||
| MNC-00114-1 | AK142231 | Rassf8 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 | ENSMUSG00000030259 | 78,19 ± 5,50 | ||
| MNC-00150-1 | AK020372 | 77,46 ± 9,02 | |||||
| MNC-00174-1 | AK016544 | 76,03 ± 15,08 | |||||
| MNC-00103-1 | AK002390 | 2700038G22Rik | RIKEN cDNA 2700038G22 gene | ENSMUSG00000086802 | NR_045042.1 NR_045040.1 | 75,45 ± 7,92 | |
| MNC-00127-1 | AK010004 | NR_027965.1 | 72,24 ± 1,63 | ||||
| MNC-00139-1 | AK131592 | 71,97 ± 13,21 | |||||
| MNC-00163-1 | AK014628 | 69,68 ± 6,78 | |||||
| MNC-00104-1 | AK018340 | NM_001242365.2 NM_001242364.2 NM_001242363.2 NM_199009.3 | 68,54 ± 14,96 | ||||
| MNC-00105-1 | AK008706 | 67,82 ± 5,18 | |||||
| MNC-00190-1 | AK007908 | 66,83 ± 6,30 | |||||
| MNC-00192-1 | AK018518 | 9030418K01Rik | RIKEN cDNA 9030418K01 gene | ENSMUSG00000070476 | 66,73 ± 18,92 | ||
| MNC-00145-1 | AK004173 | Trmt61b | tRNA methyltransferase 61 homolog B (S. cerevisiae) | ENSMUSG00000085492 | 66,58 ± 20,09 | ||
| MNC-00169-1 | AK018410 | Cdc73 | cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | ENSMUSG00000026361 | 66,42 ± 11,15 | ||
| MNC-00134-1 | AK014714 | 4833417C18Rik | RIKEN cDNA 4833417C18 gene | ENSMUSG00000086015 | NR_045187.1 | 66,05 ± 16,15 | |
| MNC-00386-1 | AK156552 | 65,81 ± 8,13 | |||||
| MNC-00232-1 | AK015544 | 63,48 ± 7,82 | |||||
| MNC-00210-1 | AK002866 | NR_040757.1 | 62,36 ± 4,60 | ||||
| MNC-00222-1 | AK020812 | NR_027894.1 NR_015487.1 | 61,58 ± 8,96 |