esiRNA TECHNOLOGY
Introduction
RNA interference (RNAi) is a powerful tool for loss-of-gene-function studies via degradation of specific mRNA transcripts. RNAi is induced by double-stranded (ds)RNAs, which are complementary to the target transcript. Introduced into cells, these dsRNAs are bound by the RNA-induced silencing complex (RISC) and separated into single strands, one of which remains bound by RISC. This strand serves as the template to recognize the target mRNA. Once the target mRNA is recognized, the protein Argonaute cleaves the mRNA and initiates its further degradation. Without the mRNA, the encoded protein cannot be replenished and will disappear. The maximum level of mRNA depletion can usually be observed 12-36 h post transfection.
In contrast, the maximum level of protein depletion is dependent on a variety of different factors, e. g. protein stability, turn-over rate, and proliferation. Usually, a significant protein depletion is detectable 72 h post transfection and may last for 48 h or longer.
Endoribonuclease prepared siRNAs (esiRNAs) are complex pools of siRNAs, which are produced from a long dsRNA molecule by digestion with an RNaseIII enzyme. This endonucleolytic cleavage mimics the digestion by the protein Dicer, the first step of the RNAi pathway under physiological conditions. Therefore, esiRNAs are the most physiological RNAi mediators commercially available. Benefit from the low toxicity, minimum off-target effects and high silencing-efficacy of esiRNAs. Confirmed by scientists from all over the world in over 200 publications.
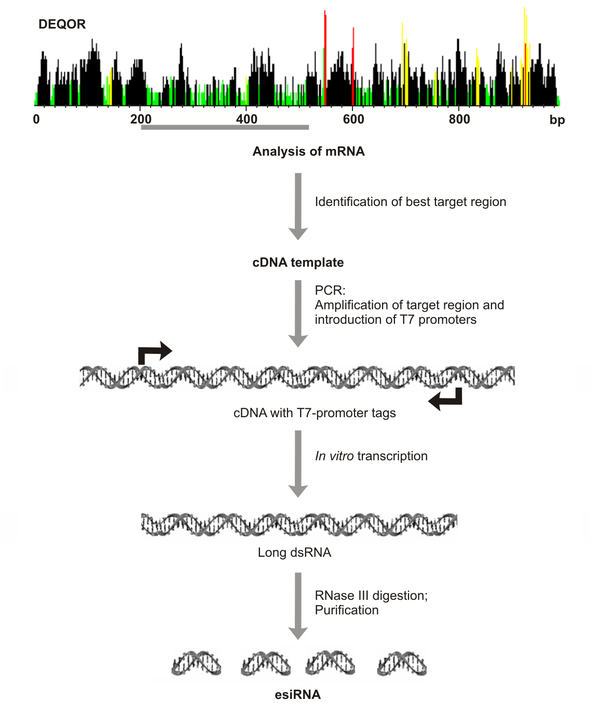
esiRNA production
The production of esiRNAs begins with the careful analysis of the target transcript-sequence utilizing the DEQOR algorithm (Nucleic Acids Research 32, W113-120). This well-established design algorithm determines the most suitable target region using state-of-the-art empirically proven design-constraints for maximum knockdown efficacy. All regions with complete or partial homologies to other genes in the respective species are avoided to ensure highest specificity. These DEQOR-predicted target regions (with a typical length of 300-600 bp) are amplified from a cDNA template by PCR utilizing two T7-promoter sequence tags. All PCR products used for the preparation of an esiRNA are verified by sequencing and assessed for their purity by electrophoresis. This procedure ensures that esiRNAs available from Eupheria Biotech are only targeting transcripts that are expressed and properly annotated. T7 RNA polymerase is then used to generate long dsRNA which is complementary to the target cDNA. This dsRNA is subsequently digested with RNase III to generate short overlapping fragments of short interfering (si)RNAs with a length between 18-25 base pairs (see Figure right). All esiRNAs are purified by anion-exchange chromatography and normalized to a given concentration, ready for transfection of your cells of interest.